Fast parallel proteolysis
Fast parallel proteolysis (FASTpp) is a method to determine the thermostability of proteins by measuring which fraction of protein resists rapid proteolytic digestion.
History and background
Proteolysis is widely used in biochemistry and cell biology to probe protein structure. In "limited trypsin proteolysis", low amounts of protease digest both folded and unfolded protein but at largely different rates: unstructured proteins are cut more rapidly, while structured proteins are cut at a slower rate (sometimes by orders of magnitude). Recently, several other assays of protein stability based on proteolysis have been proposed, exploiting other proteases with high specificity for cleaving unfolded proteins. These include Pulse Proteolysis, Proteolytic Scanning Calorimetry and FASTpp.
How it works
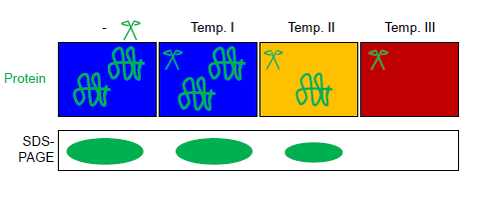
FASTpp measures the quantity of protein that resists digestion under various conditions. To this end, a thermostable protease is used, which cleaves specifically at exposed hydrophobic residues. The FASTpp assay combines the thermal unfolding, specificity of a thermostable protease for the unfolded fraction with the separation power of SDS-PAGE. Due to this combination, FASTpp can detect changes in the fraction folded over a large physico-chemical range of conditions including temperatures up to 85 °C, pH 6–9, presence or absence of the whole proteome. Applications range from biotechnology to study of point mutations and ligand binding assays.
Applications
FASTpp has been used to probe:
- Lysate effect on protein stability
- Thermal proteome stability
- Coupled folding and binding
- Ligand effects on fraction folded & stability
- Effects of mutations on fraction folded & stability (e.g. point mutation/missense mutations)
- Kinetic protein stability
Technology
First, a cell lysate is generated by glass beads beating, pressure homogenisation or chemical or physical lysis methods that do not denature the protein(s) of interest. (Optionally for targeted analysis) a protein of interest is purified out of this lysate by affinity methods based on intrinsically disordered tags or other suitable purification strategies, often involving several orthogonal chromatographic steps.
This (total or purified) protein solution is aliquoted into several tubes of a PCR strip. All aliquots are exposed in parallel in a thermal gradient PCR cycler to different maximal temperatures in presence of the thermostable protease thermolysin (see figure). Automated temperature control is achieved in a thermal gradient cycler (commonly used for PCRs). Reaction products can be separated by SDS-PAGE or western blot. The protease thermolysin can be fully inactivated by EDTA. This feature of thermolysin makes FASTpp compatible with subsequent trypsin digestion e.g. for mass spectrometry.